A second experiment to learn about the accuracy of iNaturalist observations
Today we’ve launched our second Observation Accuracy Experiment (v0.2). Thanks to everyone helping us conduct these experiments. We're learning a lot about iNaturalist observation accuracy and how to improve it.
Changes in this Experiment
We made two changes to the experimental design from v0.1 based on feedback:
- We changed the validator criteria to be at least 3 improving identifications of the taxon from the same continent since many reported not feeling comfortable identifying taxa outside of their regions of expertise.
- We messaged candidate validators rather than emailed them since many reported not noticing emails. We also only left a 4 day interval (rather than 2 weeks) between contacting validators and the deadline since last time most validating happened within the first couple days after contacting candidate validators.
Eventually, we’d like to increase the sample size from 1,000 to 10,000, but we’re sticking with 1,000 until we get a few more kinks out of the methods. The page for this experiment is already live and the stats will update once a day until the validator deadline at the end of the month, but you won’t be able to drill into the bars to see the sample observations or the validators until the deadline has passed.
New Data Quality Assessment condition for photos unrelated to a single subject
We also made one change to iNaturalist functionality in response to findings from the study. We added a new “Evidence related to a single subject” condition to the Data Quality Assessment table to make it easier to remove observations with multiple photographs of unrelated subjects from the verifiable pool.

Two of the incorrect Research Grade observations in Experiment v0.1 were of this type which we estimate to be ~350k observations in the entire iNaturalist dataset. The norm up until now to make these observations casual has been to set an identification to the nearest taxonomic node shared by the multiple subjects and then vote no to “Based on the evidence can the Community Taxon still be confirmed or improved?”, but many found this process to be clunky and confusing. We hope this new Data Quality Assessment condition will make it easier for the community to remove these observations from the verifiable pool where they negatively impact data quality and distort features on iNaturalist (such computer vision model training and the browse photo tool) that assume observation photos all relate a single labeled subject.
Thank you!
Thank you to everyone contacted as a candidate validator for participating in this experiment. We expect that considering location may decrease the percentage of samples validated compared to the previous experiment by constraining the candidates available to validate, so we very much appreciate your participation in helping get as much of this sample validated by the end of the month as possible. As always, please share any feedback or thoughts you may have on the topic of data quality. We’re excited to continue learning from these experiments and your feedback about data quality on iNaturalist and what changes we can make to improve it!
Results (added 2/29/2024)
Thanks everyone for participating in this 2nd experiment. The validator deadline has now passed, meaning that on the experiment page the stats will no longer update, the validators are now visible, and the “Accuracy results by subset” bar graphs are now clickable allowing you to drill in to see the observations behind the graphs.
In this second experiment, we estimated the accuracy of the iNaturalist Research Grade observation dataset to be 97% correct and the accuracy of the Needs ID subset to be 79% correct. The graph below shows the first experiment (v0.1) in lighter bars and this experiment (v0.2) in darker bars. The results are very close which is reassuring.

These estimates of average accuracy of the entire Research Grade observation dataset are in line with our expectations, largely because the iNaturalist dataset is skewed towards a relatively small number of common, easy to identify species (e.g. mallards, monarchs, etc.) that have an outsized impact on the average. Nonetheless, we wanted to touch on three sources of uncertainty in these estimates: validator skill, sample size, and the uncertain category.
Validator skill
We are assuming that candidate validators can perfectly validate observations as Correct, Incorrect, or Uncertain. We know this assumption is not exactly correct because there are a small fraction of situations where more than one validator looked at the same observation and they disagreed (e.g. validator 1 says Taricha torosa and validator 2 says Taricha granulosa OR validator 2 says Taricha because you can’t rule out Taricha granulosa). This happened on 1.6% of the time in v0.1 and 1.2% of the time in v0.2. This error might be higher if we are underestimating disagreements because validations aren’t done blind (i.e. validators can see each other’s validations). But the error might also be lower because each observation was validated an average of 4 times, so assuming the validations are mostly independent even if one validator made a mistake it was reviewed an average of 3 more times. In future experiments, we’ll do more work to estimate uncertainty in the labels (Correct, Incorrect, or Uncertain) stemming from imperfect validator skill. But while this uncertainty stemming from validator skill is non-zero, it’s likely close to 0. Furthermore, there’s no reason to assume that this uncertainty would bias towards inflating the accuracy by overestimating the proportion correct, it could just as well bias towards underestimating the proportion correct.
Sample size
Because being correct or not is like a coin-flip, we can put confidence intervals on our estimates of average accuracy based on the sample size. As the sample size increases the confidence intervals become narrower. We can compare our estimates and confidence intervals from v0.1 to our estimates and 95% confidence intervals if we pool v0.1 and v0.2 together effectively doubling our sample sizes.
We already have a large enough sample size (n) to have fairly confident estimates for large subsets of the iNaturalist database such as the Research Grade (RG) Accuracy Estimate. After v0.1 (n=534) our estimate and 95% confidence interval was 0.95 (0.93 - 0.96) and pooling v0.1 and v0.2 (n=1109) it is now 0.96 (0.94 - 0.97).
However, for smaller subsets such as RG Fungi our confidence intervals are still quite large. After v0.1 (n=6) our estimate was 0.83 (0.36 - 1.00) and pooling v0.1 and v0.2 (n=19) it is still 0.95 (0.74 - 1.00) - so somewhere between 74 and 100% accurate.
For very small subsets, our sample size is still much too small to provide useful estimates. For example, to estimate the accuracy of RG Rare (taxa with fewer than 1000 observations) African Insects our estimate even after pooling v0.1 and v0.2 (n=5) is 0.6 (0.15 - 0.95). For other subsets (e.g. RG Very Rare (<100 obs) African Insects) we have a sample size of zero and can’t make any estimate.

The size of our sample is adequate for getting relatively confident estimates of average accuracy for the entire iNaturalist database and for large subsets (e.g. the RG subset, the North American subset, the Insect subset etc.), but these sample sizes are too small to yield confident estimates for more niche subsets (e.g. RG South American Fungi from 2022, etc.). We are very interested in variation in average accuracies across these subsets and look forward to growing the sample size to the point where we can better understand this variation.
Uncertain Category
Ideally, we’d be able to label all observations as Correct or Incorrect. But because we don’t have the capacity to get validations on all observations in the sample, some remain Uncertain. This was 3% of the RG subset in each of v0.1 and v0.2. Since we are calculating Accuracy as the percent correct (as opposed to 1 minus the percent incorrect) this Uncertain category is biasing us to underestimate the Accuracy. The true average accuracy of the iNaturalist Research Grade observation dataset is somewhere from 0% to 3% higher than our estimates because of this bias which is on par with the uncertainty interval resulting from sample size.
Thank you and next steps
Thanks again for all your help with another successful experiment. We’re amazed by the capacity of this incredible community to validate these samples. We hope you’ll click on the graphs and explore the results here. We plan to run another experiment at the end of March. We may try keeping the methods the same and increasing the sample size from 1000 to 10,000. Or we may make another change to the methods such as more changes validator candidate criteria. Thanks again for making these experiments possible!





Comments
Thank you for your work!
Happy to have provided my 2 IDs.
Even happier about the new 'single subject' feature!
Great ! More of this please :)
The new DQA is great, but I think "at least 3 improving identifications of the taxon from the same continent" is not a criterion that is likely to lead to accurate data for 2 reasons:
1: Requiring the improving IDs to be on that exact species means that even the world's top expert on a rare species may not count as a validator for that species if the species is so rare that no one person has made 3 confirming IDs (eg there are 10 obs IDed by 9 people)
2: Three is too small a number to rule out people who don't know what they are doing yet
I think it should be "at least X number of improving identifications of genus level or lower taxa in the same family from the same continent" and X should be a 2-4 digit number
About 1: Rare species are rare, so the impact on the data will be small. I'd be willing to bet there is not a single such case in the 1000 obeservations.
Any one rare species will not have much impact, but when you add up all the obs of all the thousands of rare species it could be meaningful
Done. What a mess of non-flowering, probably unidentifiable dicots. But I put a label (e.g., dicots) on everything so you'd know I participated.
'identification to the nearest taxonomic node shared by the multiple subjects'
does this still apply ?
I understand this is not required anymore.
"The page for this experiment is already live" has a broken link to the the staging environment. It should link to https://www.inaturalist.org/observation_accuracy_experiments/3
So great to have some resolution on the "Single Subject" issue. Thanks.
But what do I do with "duplicates"? Observations that are obviously of the same organism at the same place and time, but spread over several observations, or even a straight duplicate observation using the same picture for the same organism.
For this data quality assessment I should post the finest ID I could, but that would count as an improvement, on an observation others have looked at previously and refused to ID (as I would have) because it was a duplicate, and for which a much finer ID already exists on the original.
So by posting my ID I will artificially improve the identification (from Tribe rank to species), which is already at the species level on the original observation. And in doing so I will support a practice that I am totally against, and state so on any observations I encounter (and for which I have been censured for identifying them as "Life" whenever I encounter them).
So, if you dont mind, I shall ignore this observation. [And not disclose it so as to limit other fanatics from trying to swing it one way or another. ]
new DQA??? I look forward to using it. and to bug reports...
sorry - staging link updated
Sorry - updated to /44
should be /3
I like the changes to the experiment design. But I do share the concern that "at least 3 improving identifications of the taxon" might not prove sufficient expertise to validate IDs. Narrowing things down by continent is definitely a huge improvement but many identifiers are much more specialized on a certain region. Asking someone who usually identifies observations from Alaska to identify an observation from Florida might not yield optimal results. But I am very glad that these experiments are done, and I find the results very encouraging!
Thanks. It is very nice to get a set of observations that I can actually identify, rather than from parts of the world where I am clueless.
By the by, one issue that might skew this experiment is getting notification of disagreements by other identifiers as part of this experiment. (Not that I erred, but the duplicate mentioned above has generated 2 identifications since I posted a comment deferring ID on it less than an hour ago).
And I am guilty of pointing out to another participant that he was using an old concept ID, and that the species that he had agreed to had been split. However, it seems to me that such is how IDs get improved normally on iNat, so this is just fastracking the process. It does however mean that the IDs are thus not independent, but influenced within the experiment. I dont know if this matters to the design assumptions for the experiment.
Echoing the love for the new “Evidence related to a single subject” condition to the Data Quality Assessment!
What is perhaps missing from the experiment is "Improved" (we currently have Correct, Uncertain and Incorrect, leading to Accuracy, and a second index of Precision [presumably calculated on the final result, after validation])
At least two of my sample where above species level and could thus have been improved [one already has improved from Tribe to species). (another one was at species but had subspecies, and so could also have been improved - 2 others were already at subspecies ID at the start of the experiment, but this experiment does not consider IDs below species rank).
Surely the fact that observations can be improved is relevant to the accuracy of the identifications on iNaturalist? In terms of https://www.inaturalist.org/observation_accuracy_experiments/3?tab=methods
what is relevant is observations for which the Precision has improved. (or more specifically: Correct Observations for which precision has improved).
The method would be to compare the Precision at the start of the experiment with that at the end (for correct observations).
Or two paraphrase: "Are the correctly identified observations on iNaturalist as good as they can be?"
In the 'Methods' tab of the Observation Accuracy Experiment page: the figure of 'Step 1' lacks an explanation about what the brown 'C' hexagons are.
I am rather unreasonably pleased about the new DQA functionality! Thank you. I shall continue to ID to the nearest common rank until it is said otherwise. (If only because that's sometimes what really alerts people to the fact they need to do something, which a DQA won't.) Duplicates are the next hurdle as mentioned above, but it's good to see things being worked on bit by bit :)
I agree with @tonyrebelo that seeing other identifiers IDs and comments will mean the the results based on subsequent IDers are essentially uncontrolled/not independent. On one hand, having IDers be able to use others comments/IDs mimics the process of IDing on iNat and is realistic. However, because IDers have information other than that present in the evidence (ie, photo), they will use it when making their IDs, so a rigorous statistical analysis of any data generated by this protocol would be difficult/impossible.
I am really excited about the new DQA! I think this will give a quick fix to a lot of observations that aren't ideal and free up identifier time and effort for other things. It would be great to have a recommended workflow for multi-taxa observations with the new DQA as @dianastuder asked about. Based on my initial understanding, using the DQA (eg, a downvote) could be done with no other action and be legitimate. An IDer could also still ID to the lowest common taxon which might be helpful in some cases (eg, two species in the same genus), though this isn't necessary. The IDer could also leave a comment asking the observer to delete/split photos and leave a comment when they do so (since these wouldn't generate notifications), though again, this wouldn't be necessary. Observers often don't respond to requests like this left in comments, but sometimes they do, and, in these cases, the DQA vote could be withdrawn if an observation is corrected to focus on a single taxon. Any other thoughts about best practices for using this DQA?
I think making a coarse ID is still optimal given the photo browser page for each taxon includes images from casual observations by default. So only adding the DQA vote wouldn't solve one of the problems the new DQA was designed to address, ie images of wrong species in the wrong photos page
I'm also very glad to see the new DQA flag. I'm looking forward to a blog post (or better still an addition to the help docs) explaining how identifiers should use it. In the meantime, I'm intending to add it to my standard process for "multi-subject" observations.
ID at the lowest level that covers all photos.
Add a comment explaining how to split observations and tag the user.
Downvote "Evidence related to a single subject"
Also... is it possible to query the new DQA field via a search URL or via the API?
Love the new DQA and that the experiments are going on. 👍
Hi all - thanks for all the engagement and great feedback on these experiments. We updated the stats (we’re doing this manually once a day now but will automate this in future experiments) and are already up to 548 (39%) of candidate validators with 90% of the sample validated. So exciting to see the capacity of this awesome community to do these kinds of experiments. Thanks so much!
@sqfp thanks for catching that error, a fix is in process. We’ve tried to bin the feedback raised so far:
Validator criteria
Lots of great ideas on how to improve validator criteria:
__ @insectobserver123 proposed loosening constraints on taxonomic criteria: use ancestor taxa (e.g. family/genus) rather than species (e.g. >3 improving ID of the genus encompassing this species)
__ @insectobserver123 proposed further constraining number of improving IDs criteria (e.g. increase from >3 to >10+)
__ @matthias22 proposed further constraining geographic criteria: from continent to state
As was raised, the strictness of the validator criteria trades off with our capacity to get samples validated, so it’s very interesting to keep an eye on this as we learn more about what kind of response we’re getting from the candidate validators (which again so far is very heartening). Re: @richyfourtytwo’s point about rare species, the 1-100 bar of the Taxon Observations Count graph is a good proxy for rare species. If you toggle the show frequency button you can see that it represents 35/1000=3.5% of the random sample
Norms for responding to observations of multiple species
Echoing ideas raised by @thebeachcomber, @dianastuder, @richyfourtytwo, @matthewvosper, @cthawley, and @rupertclayton please prioritize (1) voting no to the new DQA condition. After that it’s also helpful to (2) add IDs of the nearest shared taxonomic node and (3) to politely ask the observer to split the observation. But since 1 will move these observations to Casual, 2 & 3 are not as high priority if you don’t feel like doing these additional steps. Please don’t vote no on the “Based on the evidence can the Community Taxon still be confirmed or improved?” question for multispecies observations since we can now disentangle those. It’s not possible at the moment to query observations by DQA metrics though DQA metrics are contained in the observation API response.
Duplicates
@tonyrebelo and @matthewvosper both raised concerns about Duplicates. We agree that duplicates can complicate assumptions about the sampling bias and the independence of observations necessary for certain statistical analyses- and they can also be frustrating for IDers by creating extra work. But we’ll have to think more about how they are related to this framework of improving Accuracy and Precision. They are certainly related if they contribute to IDer fatigue or frustration. To be continued, but in the meantime, please don’t change your ID behavior on duplicates, even if you justifiably find them frustrating.
Other things we could measure
@tonyrebelo and @cthawley brought up concerns about validators being able to see existing IDs (e.g. this not being a blind assessment). Under the assumption that our validator criteria is sufficient, the fact that validators are no blind won’t impact our estimates of Accuracy and Precision of the sample of observations. But we agree that it would be very important to do a blind experiment if we were trying to measure validator skill. And that this might be necessary to test our assumptions about validator criteria mentioned above, or to learn more about related topics such as identifier skill.
@tonyrebelo also raised the idea of measuring the related question “Are obs as finely identified as they can be?”. We agree this would be a very interesting thing to measure, but would require different validator criteria and a different experimental design from the experiment which is meant to answer how accurate and precise the observations are at the time the sample was generated. But we very much agree that it would be interesting to measure what precision is possible, because as is implied species/subspecies level precision isn’t possible for many observations.
Thanks again and keep the feedback coming, we'll update the stats again tomorrow.
Excellent point on blind assessment by @tonyrebelo and @cthawley! I think this should definitely be done in some future iteration of the validation experiment.
@loarie one point I should probably make is that the stricter the other criteria are the smaller the ID number threshold should be, I was thinking of anywhere in the 50-2,000 range using family and continent, but using genus and state the number should be smaller to avoid making the criteria too strict and to avoid the rare species issue, though I still think 3 is too small for any scenario. I also think genus is likely too fine a taxon, some genera are like 1-2 species. I also would suggest country as a geographic measure to consider, in addition to state and continent
Since this is a scientic experiment, I'll give an objective assessment, whether it will end up being mostly agreement, disagreement, or a mixture, depending on the nature of the experiment. First, for at least one positive note: most of us are at least willing to participate in this even if we think it has too many design flaws or limitations, just because we like to ID. Starting with commenting again on the most recent experiment, Scott announced it apparently without consulting many (if any) non-staff identifiers upfront, which prevents the community from usefully correcting design imperfections. As identifiers commented on both of his announcements for the earlier experiment, the threshold/standard for being a validator remains far too low. Someone merely guessing IDs could easily achieve it.
We messaged candidate validators rather than emailed them
For users who staff typically give silent treatment to in direct messages or emails (or tags, etc.), are they still counted as validators in the experiment and results if they make the instructed IDs but don't reply to the message? Another question: presumably this experiment is also based on RG accuracy (?), but RG seems to barely if at all be mentioned in this announcement.
Eventually, we’d like to increase the sample size from 1,000 to 10,000, but we’re sticking with 1,000 until we get a few more kinks out of the methods
Both numbers are actually insufficient sample sizes for what the experiment's trying to achieve. So, the results shouldn't be reported as if they're highly confident or accurate estimations, as they were last time. And it would be very misleading to publish them in external sources - since later more accurate experiments will likely contradict past claims like the 95% RG accuracy. Recently, some of the Bug Guide contributing editors (curators/experts) thought the 95% inat claim was accurate, until I explained the experimental flaws and that the community input wasn't effectively incorporated, resulting in them correctly concluding that the 95% was "a stretch" (a big overestimate, and the true accuracy isn't even known, but we do have reason to know that BG's is far higher).
We also made one change to iNaturalist functionality in response to findings from the study. We added a new “Evidence related to a single subject” condition to the Data Quality Assessment table to make it easier to remove observations with multiple photographs of unrelated subjects from the verifiable pool.
Yet, a further better feature that BG has would be even better: to allow such obs. to be "unlinked," generating two "child" obs., one per species, that remain verifiable, as many of us have long suggested. Incidentally, the fact that it took 5+ years for even this current DQA option to be added was part of the reason that two non-staff curators flagged me a year ago, which was part of why Scott invalidly removed my curator status. Talk about people now having an "idea" people who previously had were punished for in the past. Secondly, "evidence related to 1 organism" is a confusing description, that will inevitably lead many users to uninentionally misuse it (like so much else of the DQA and other website's obscure wording and terminology).
this type which we estimate to be ~350k observations in the entire iNaturalist dataset.
Which was apparently okay to happen for 5+ years prior. As the bee researcher LeBuhn once wrote (paraphrasing), species IDs are all that really matter (excepting few genus data studies). The previous inat requirement to use broad IDs for multi-species obs. (which many identifiers, including some admin, justifiably "disobeyed") rendered 350,000 would-be species-rank records nearly useless. My point isn't to criticize Scott or the idea of these experiments (we want to test accuracy), but to point out that he essentially took very little of the experienced identifier feedback on the last experiment into account, and that many problems remain. Turning now to reply to some of the commenter comments:
But what do I do with "duplicates"? Observations that are obviously of the same organism at the same place and time, but spread over several observations, or even a straight duplicate observation using the same picture for the same organism.
Exactly.
I agree ... that seeing other identifiers IDs and comments will mean the the results based on subsequent IDers are essentially uncontrolled/not independent
Yes, but not every aspect of any experiment can be perfect, and as the inat admin have always claimed, there are so many constraints to what they can and can't do. Instead, I'd suggest focusing mostly on the major methodological flaws (e.g., too small sample size, too low validator threshold, etc.).
@bdagley
The problem with this discombobulation is the assumption that all pictures were taken at the same place at the same time on the same day. That is certainly not valid. Where location and date are in the exif, they could be used to inform the observation, but geotagging photos is not the default, and even most apps dont do it.
So it is not a simple, straightforward process.
I would support any improvements that happen to the system, but you have to do something about blurry photos. I see some users uploading blurry, unrecognizable species and receive identifications. Personally I wouldn't feel comfortable identifying a species from a blurry photo but it seems to be happening. I would really like to see inat doing something about this as well.
Since the new DQA is 'hidden' in this post - with a quite different title - I add a comment when I use it
new DQA Feb 2024 - only one species please
https://www.inaturalist.org/blog/90263
And this needs updating please ?
https://help.inaturalist.org/en/support/solutions/articles/151000171680-what-do-i-do-if-the-observation-has-multiple-photos-depicting-different-species-
@marieta55, identifiers should not add identifications that they are not confident in. But I'm not sure there's much we can do to prevent identifiers from ignoring these guidelines. We can detect blurry photos, but they don't necessarily indicate that observations are unidentifiable. For example, compared to possible alternatives I'm confident this is Chinese Pond Heron based on the breeding plumage pattern and that this is Giant African Land Snail based on the shape despite both being extremely blurry photos.
If an observation is sitting at taxon X and you are confident other taxa can't be ruled out because the photo is too blurry, please add a disagreeing Identification of the nearest ancestor you're confident in and explain in a remark why you don't think the evidence supports ruling out the alternative(s).
@marieta55 blurry photos.
A photo does not have to be in focus for diagnostic features to be clearly visible. Photos out of focus are not a reason not to make an ID.
If the photo is out of focus and no diagnostics are visible and ID is not possible - and you are certain that others who are more comfortable about identifying species from blurry photographs cannot make any ID from the photograph - then why not use the option "Evidence of organism" = false?
But surely one can make a more general ID to "insect" or "plant" to the level that you are comfortable with.
Nice hearing you are always improving!
Good improvement.
@tonyrebelo I didn't mean slightly out of focus photos. I am talking about shades and shapes like the following link: https://www.inaturalist.org/observations/191483453
This particular user seems to be quite knowledgeable about birds but I believe that he doesn't have the right gear to record his observations. The majority of his observations though seem to be of poor quality, zoomed in to pixel level which makes them unrecognizable. Personally I believe there should be a standard for acceptable photos. Of course I could be wrong but I do take a lot of time trying to improve the quality of my photos so that my contributions can be usable to people.
@loarie I agree with you. If I don't feel comfortable confirming an id, I don't do it.
The observations I got were of species I wasn't super familiar with. One was a Sarracenia in a California botanical garden, but the only ID's of Sarracenia I have are of adding subspecies level ID's to Sarracenia purpurea in Ohio, since there's only one subspecies present here. The other observation I got was of an Echeveria flower (no leaves), but in the Crassulaceae family I usually only ID Kalanchoe so I'm not very good with identification of the rest of the family.
I'm curious why you all are conducting these experiments when you already have the data needed to assess the accuracy of iNat IDs. The "Taxa Info --> Similar Species" page lists all the misidentifications for each species. It seems to me, you'd be better off performing analysis of that data.
I've analyzed this data for 46 species I've been following on iNat, and many of the misidentification percentages are over 30%. So forgive me if I don't believe your estimates of 95% accuracy for all iNat IDs. It seems to me the accuracy of identifications depends on species, and all species shouldn't be thrown into the same bucket when calculating and reporting the accuracy of iNat data as a whole.
It seems to me that your experiments are analyzing the process of identification rather than the accuracy of identification for all the reasons stated in the comments above.
Misidentification of Erodium observations in CA - Updated
Misidentification of Geranium Observations in CA - Updated
Misidentification of Robertium observations in CA - Updated
Misidentification of Malva observations in California
Misidentification of Fumaria Observations - USA
@truthseqr I think everyone knows that accuracy is dependant on taxon - that's why it was made possible to break it down at least a little bit. The sample size of these initial experiments is too small to dig down meaningfully too far though. It would be a mistake to do a huge experiment straight away. Most iNat observations are not of very rare and difficult taxa, so it makes sense that accuracy per observation will be greater than accuracy per taxon.
The similar species page measures something totally different - it records every disagreement with an ID of that taxon, not just disagreements after reaching research grade.
Single subject rule violation has two common causes:
1) Fat fingers - observer adds photo to the wrong observation. Most of these people are prolific observers with a lot of experience.
So far found ~ 15, left note and all were fixed.
I done it 5 times when uploading multiple observations and dropping the photo to the wrong one . I got comments and cleaned them up.
2) Multiple, similar looking objects
Half a dozen example so far.
I usually leave a note explaining the difference and asking for a split.
All were fixed. The longest took two weeks.
Question: Who can clear the "single subject" cross and how easy it is?
Question: Who can clear the "single subject" cross and how easy it is?
If it was your cross, you can click it off again.
If not yours, then you click a counter-vote.
Does the new DQA value cancels RG? Hopefully not! It is good for not misleading the learning artificial intelligence. But the condition of its applicatoin should be somewhere explained very clearly, as to the scenarios dicsussed before. I do not think users are instructed well when to apply this value and expect its unjustified overuse. Those scenarios were:
1) More than one organism seen in the photo (which is almost always the case), with 1a) the main object obvious for a human visitor (e.g. the butterfly among vegetation), but maybe not for a robot, or 1b) not obvoius, e.g. several tree species in a photo, with indication in the notes to which object the observation is devoted (e.g. 'this observation is for the yellow tree', or 'this observation is for the flower., not for the butterfly), or even without a note but with a certain inidtal identfication of the author;
2) some further photos deliberately show something other than the object, on the main photo e.g. its habitat , or informational banner;
3) Several species mixed, e.g. different butterflies, which the photographer incorrectly assumed to be the same.
The scenarios 1 and 2 are deliberate and justidied; such observations may be removed from the AI learning sample but not from research grade. BUT most photos do have some other organisms in view (e.g. grasses on the background), which would make application of the value very arbitrary.
To remain on the safe side, I would recommend and even insist to change the wording from
“Evidence related to a single subject”
to
"The subject is concerned in all photos"
(or something with better English)
@matthewvosper,
1) If the "Taxon Info --> Similar Species" page measures ALL disagreements with an ID of that taxon and this is not something the team wants to measure, why not create a different page that measures disagreements after reaching research grade? To do this programmatically is much more efficient than conducting many small experiments of this nature.
2) If an observation is misidentified, it could sit in the database for years before it's corrected and captured by the criterion that misidentifications are only counted when they're posted against a research grade observation.
3) I would suggest that the team should not proclaim 95% overall accuracy as was done after the first experiment, given the fact that misidentifications are species-specific.
4) I have personally viewed thousands of misidentifications of Geranium, Erodium, Fumaria, Malva, and other genera. That's a lot of misidentifications and it makes me skeptical of claims of 95% accuracy.
5) The reviewers are vetted for these experiments, which is not the usual case for iNat reviewers. I think this skews the data considerably.
@oleg_kosterin, thanks for pointing that out - our intention was that
We'll work on getting this into the help documentation
@truthseqr I'm a huge fan of your misidentification journal posts. Thanks so much for continuing to do all those studies.
As matthewvosper mentioned, these Observation Accuracy Experiments are currently designed to estimate the overall accuracy and precision of all iNaturalist observations. In contrast, you're measuring the rate of misidentifications which is also very interesting and absolutely something we should learn more about as it relates to identifier skill. But they are different things. For example, for Fumaria densiflora (choosing Filter by Place = United States) I see 68 observations and 6 records of Fumaria officinalis under the Similar Species tab. But from an Observation Accuracy standpoint, none of those observations are currently not Accurate since they've all been rolled back to genus (we're measuring the difference between an obs sitting at Species and an obs sitting at genus as lower Precision but identical Accuracy).
As mentioned we're aware the current sample size of 1000 might be too small to provide robust estimates. But Observation Accuracy Experiment v0.1 estimated the accuracy of the Research Grade subset to be 95%. The accuracy of the Verifiable subset (which includes Needs ID as do the counts on the taxon pages) estimated the accuracy to be 90%
But the big difference is your studies are measuring the misID rate, and these studies are measuring the accuracy of observations which takes into account things like the observations rolling back to genus when there are disagreements. In terms of measuring misID rate, I think the percentages are actually worse than your studies estimate because the Similar Species tab only detects situations where alternate IDs were added, not all the observations with just 1 misID where alternate IDs have not yet been added.
@loarie, thanks for explaining all this to me. I wish you much success in your efforts to determine the accuracy & precision of iNat data.
@loarie, thank you for the explanation. Yet the issue if this value affects research grade or not is important - those controversial practices like habitat photos or bat sonograms are undesirable for AI learning but helpful in various biological respects, including those desiranle in GBIF, which adopts only RG observations. I hope these uses can be separated at the software level, like making somewhat different subsets of data passing to such general data using/accumulating systems like automated identification and GBIF.
Suggestion for duplicates: provide a checkbox to identify an observation as a duplicate so at some future date we don't have to go back and search for all the duplicates.
Not sure if I am out of topic here, but I have noticed people are uploading images that are tempted. For instance, a person draw on the image, trying to be funny. I have reported the image, but the moderator said it was not an issue. Maybe, a box for separate those images, where the observation was modified, like any draw, from the pure images. It is useful for machine learning to use just pure image.
The meachine learning should learn what iNat users are uploading, which may include stuff like arrows etc. So such data may actually be useful. Of course I don't know what drawings you are referring to, probably something else, bacuase arrows are hardly funny. :-)
for duplicates @truthseqr
I put the other obs link in a comment
and vice versa
or - list all 3 or 4.
I've seen them, I've found them - I have done the work, just need to copypasta in a comment.
@dinastuder, it looks like you have a technique for finding duplicates, but I'm not sure I understand it.
I do step #1 also, but the comment refers to the original observation that I want to keep, not the duplicate observation that needs to be deleted (e.g., "This is a duplicate of xxx. Please delete it.")
I don't know what you mean by "or - list all 3 or 4".
I've used the "Duplicates Observation" field, but again it refers to the original observation, not the duplicate that needs to be deleted.
If there's a way to search my comments on other people's observations, then I'm not aware of it.
@richyfourtytwo I am saying to have a tag, as so we can decide what to feed our models. With a tag, we could use a filter to filter out those images.
Adding a few more notes re: the experiment design, and replying to comments. I found the very long description of the last RG experiment somewhat confusing to understand/read. For the current experiment, the description was concise, but left questions, at least before clarifying comments were made. E.g., it says "We’ll calculate accuracy by comparing your ID to the Obs. Taxon," but RG isn't mentioned. So, it was at least initially unclear if that meant that only RG, or RG and Needs ID, or RG, Needs, and casual accuracy are being calculated. By inference, "ID all obs. in the link below as finely as you can" would imply that some obs. will end up being RG, others Needs ID, and others casual.
@tonyrebelo The problem with this discombobulation is the assumption that all pictures were taken at the same place at the same time on the same day. (re: me suggesting that obs. with 2 species should be split into 2 separate obs. as Bugguide.net does)
In such situations, the original obs. has both photos, which typically are taken on the same date and place, or the observer can be asked/assessed. It may also be possible to look into the photo metadata to tell (?). But, if there was an obs. where it's truly unlikely that the photos were from the same date and location, they can still be split/unlinked, and just marked "location or date inaccurate" in DQA (rendering them casual). The reason why Scott's current DQA proposal could improve to be like that is because his DQA vote makes the entire obs. photos casual, vs. "my suggestion" (which is actually from BG, where it works well) saves at least some verifiable records. Vs., in the new current system, even some rare species sightings or new locality records could become casual (lost).
@marieta55 citing the "problem of blurry photos" and referring to obs..
We'd ID these obs. the same way as usual, to the finest level they can be. If a photo seems not to show wildlife, that means making it causal via DQA "no evidence of organism." In the obs. e.g. you gave, it does clearly seem to be a bird, but would be justified to ID at a broader bird rank.
@truthseqr So forgive me if I don't believe your estimates of 98% [correction 95%] accuracy for all iNat IDs.
While I didn't check your method for calculating this, of course the previously claimed 95% RG accuracy estimate from the last experiment was a high overestimate, and the experiment wasn't designed in a way to be able to make any meaningful estimation. Commenters explained this both in the post announcing that experiment and in the one reporting on it’s results, but their points seem to have been mostly ignored.
@oleg_kosterin I would recommend and even insist to change the wording from “Evidence related to a single subject” to "The subject is concerned in all photos" (or something with better English)"
Exactly. Something like the “subject or focal organism is in every photo.”
@dianastuder And this needs updating please? https://help.inaturalist.org/en/support/solutions/articles/151000171680-what-do-i-do-if-the-observation-has-multiple-photos-depicting-different-species-
Definitely. Incidentally (but which is on-topic) also that that page that refers to the past policy, says "it's best to identify to the level that fits all photos," which means that was technically only a suggestion, not a mandate. Yet, I was flagged (and singled out among other users who made the same ID) by two curators a year ago who mistakenly asserted that I violated site policy, which was part of why my curator status was invalidly removed.
@loarie But Obs. Accuracy Experiment v0.1 estimated the accuracy of the RG subset to be 95%. The accuracy of the Verifiable subset (which includes Needs ID as do the counts on the taxon pages) estimated the accuracy to be 90%
But we already know, from our inat experience, and many reasons users gave here and elsewhere, that both of those must be (probably high) overestimates. It's misleading to refer to those results as "estimates," which could imply high confidence. Secondly, your older link now directs to a nonexistent page. Another relevant thing to do would be to calculate the average CV accuracy across all taxa when an observer uses it as the first ID and there are no subsequent IDs added. That might not even require a user-experiment (e.g., we know that photos of grass aren’t birds). I predict/infer “initial CV accuracy” to be low. From that, we could infer more about how accurate these recent accuracy estimates truly are. Plus, as you here granted, the sample sizes are too small in the new experiments. Validator eligibility also remains far too low, which you didn’t address.
@ any/all users, and which is on-topic here, I recently compiled a list of requested improvements for all aspects of inat (some of which were from other users), which is unique in including some requests that aren't on inat or the forum. Including e.g.'s like suggesting observers crop photos, an AI auto-photo cropping tool, an AI tool to ask "is an organism present?" for photos that seem to lack any, and a request to "recommend or require observers don't put watermarks or logos on top of the animals in their obs." The last one was actually censored from being posted in the past by staff, despite that they said it wasn't inappropriate. Many of the requests, if voted on or implemented, would significantly increase the accuracy of the CV/AI and of RG % accuracy.
Since the request list is very long, being a compilation (but probably shorter than the forum feature request list), I recommend viewing only a portion of requests per day. Some requests near the end are about website fairness, privacy, etc., but are written appropriately with constructive/cooperative intent re: the admin. Feel free to share with anyone who might be interested. The requests I wrote come from 4 years of extensively using inat and BG (which has higher accuracy) and often thinking about accuracy. Thanks for considering it. https://docs.google.com/spreadsheets/d/1b0VFdSO5iI8PbovZ4YNjyoIUGxP2KOy8iFZXz-ak43I/edit#gid=1754535465
If there's a way to search my comments on other people's observations, then I'm not aware of it -@truthseqr
https://www.inaturalist.org/comments?utf8=%E2%9C%93&mine=true
then use the search box
In this discussion of why 95% accuracy must be an overestimation, I think something important is being ignored - the variation of error rates among taxa. Many species are virtually unknown except by people really interested in them, so they're rarely misidentified at the species level and any misidentifications rarely make it to RG. Also, some taxa are regularly checked and corrected by dedicated identifiers So although we're all well aware of (and frustrated by) taxa for which misidentification is common and error rates are high, we should remember that those get averaged with other taxa that have really low misID rates, especially for RG observations.
Others may discuss your comment, but as for me, I'd agree that what you pointed out could have an effect, but a somewhat small effect. For reasons I and others gave (especially in the comments on the posts about the first experiment), there are many reasons to think the 95% must be high overestimate.
Separately, I'm wondering could anyone provide a list of links to past inat accuracy tests/experiments/estimations? Even ones with different designs. The one that Scott linked to apparently no longer exists at that url. I'm also wondering if CV accuracy has ever been tested. Or, in the even that no one can link to the past similar studies, can they at least indicate the most efficient way I could find them all? I assume most or all were posted on these inat blog/journals, although that some could've also been on the forum, or elsewhere. In general, I find locating past posts like this difficult to do.
It would be great to move some of this discussion to the forum where more messaging and moderation tools are available.
Thanks all for your continued help validating sample observations. We just crunched the stats as of today and we're at:
-- Mean number of validators per sample observation: 4
-- Number of participating validators: 803 (58%)
-- Percent of sample validated: 96%
@bdagley thanks for catching the broken link - Its fixed. The links to the 2 experiments are:
-- v0.1
-- v0.2
Thanks again all!
Given that not everyone on iNat is on the forum I think it is good to be able to discuss things directly on iNat, though there is nothing wrong with talking about this on the forum as well
@loarie I was actually referring to, seeming to remember, even earlier calculations (maybe not "experiments," as I said) from years ago, possibly originally shared on the forum, that I think you or other admin once shared. In other words, I'm asking if there are links to any past calculations that had anything to do with Needs ID accuracy, RG accuracy, AI accuracy, etc. Unless there were no other ones, in which case I'd ask if what I'm remembering were past calculations about what percent of species had been observed (etc.), or something else like that.
Given that the concept of the new DQA vote option of whether the ID subject is in every photo of the obs. has come up in this discussion, and that I (as in the past) suggested that it would be even better to allow the obs. to be unlinked (split) into two verifiable obs. (vs. rendering both photos unverifiable), that terminology that some identifiers like johnascher use originates from the similar website and early inat influence Bug Guide. This is an example of how it works there: https://bugguide.net/node/view/671812.
Perhaps I'm not geek enough, but am I the only one who is still confused as to what is being termed (and measured) as "precision"? On the methods tab, the definition of "precision" starts off with: "If every sample observation was at a coarse taxon..." Could someone please add a first sentence or two, in plainer English, describing what precision is? Perhaps a simple declarative sentence that starts with, "Precision is a measure of ..." and some follow ups like, "High precision would indicate a large proportion of observations are identified to species level. Lower precision would result if more observations remain at genus or higher taxonomic level."
I have to trust that the subsequent description of how "precision" is calculated is statistically useful. I can't follow it, nor can I understand how the "51%" example is calculated. But I don't need to.
@gcwarbler lets use the 30 species of the duck genus Anas as an example.
If an observation was at species Mallard, its precision would be 100% because its precisely placed to a single species (Mallard). If the observation was at Genus Anas, because there are 30 species below the genus the Precision would be lower: 1/30=3.3%. If it was ID'd at family Anatidae, since there are 165 species below the family, the Precision would be 1/165=0.6% and so on.
Just like we report average Accuracy by taking the average of the Accuracy of each observation in the sample, we're reporting average Precision by taking the average of the Precision of each observation in the sample. For RG obs its close to 100% because aside from a few exceptions RG obs are all at species. But its lower (~77%) for Verifiable obs which aren't.

The reason we want to measure Precision is that it trades off with Accuracy. For example, if we rolled back all Duck observations to Family, the Accuracy would probably be very high, but the Precision would decrease. We want a dataset that is both Accurate and Precise.
The Precision statistic is nice in that it gives obs sitting at species a high value (100%) and obs sitting at coarser nodes lower values (approaching 0%), but its not ideal. For example some drawbacks are:
-- Its based on the number of species in the iNat taxonomy. This works well with ducks since all 165 are in the iNat taxonomy, but for groups like beetles where we probably only have a small percentage in the database and more are added manually every day, the denominator is shifting.
-- Like CV and the Species counts, we're ignoring subspecies and hybrids which isn't ideal because people do ID at these nodes. But if we included the 2 mallard subspecies which are rarely used by IDers, an ID of species Mallard would have a precision of 1/2=50%
The link you send me don't work again. It append the same in the first experience. Can someone please send me again?
@amanithor can you email help@inaturalist.org to try to troubleshoot your issue with the URL? I suspect it has something to do with your browser
Great
Given the tangential mentions of CV accuracy above I might just mention a little experiment I did with the hoverfly genus Eristalis last year and the year before. First time I sampled 100 needsID whose current ID was at genus, second time only 50. I found the CV got the species right 75% and 76% of the time respectively. This included as many as 12 species on 5 continents. I thought it was pretty good. For context, Eristalis can be considered a genus that is difficult to ID from photos, but rarely impossible. It does generally have a lot of iNat observations to train from, but not for every species. Most of the errors were from poorer quality pictures. I thought that was pretty fantastic - certainly good enough to make identifiers lives easier. Maybe I should repeat it!
Personally I am impressed enough to recommend that the AI CV automatically identifies (and posts the ID) of all "Plant" rank identifications to Family level. At this stage I am reluctant to suggest Generic level, but I suspect in a few years time the AI CV will be far more reliable than the average identifier at even species level.
All the specialists bemoaning how badly the AI CV does with rare species, should bare in mind that (1) the AI CV has not been trained on these species yet, and so does not know about them; (2) unless the specialists identify these species then the AI CV will never know about them, & (3) +95% of identifiers on iNaturalist dont know about them either, and are no better than the AI. We need to keep our expectations realistic. And we all know where the untrained species will be found - under the most similar common species: common species identifications will need to be confirmed, and rarer species correctly identified from among them. That is simply how it works - with both human and the CVAI training, and both on iNaturalist and in the field and herbarium/museum with trainee specialists.
Even (or should I say: most especially) humans are going to perform very poorly on the really localized, really sparse, seldom recorded, and not-in-popular-fieldguide species.
Does anyone know if there have been any studies on the accuracy (and precision) of identifications in the typical herbarium or museum? I suspect iNaturalist overall will perform far better on the overall collections (although hopefully the museums will perform far better in the local specialist collections, and the curators' specialities).
I agree with @matthias22 that constraining validator eligibility to continent is an improvement. However, instead of further constraining eligibility based on political boundaries, why not put a circular boundary around the observation and use that as the selection criterion?
1 of 2
@loarie These are the past accuracy experiments/calculations I remembered and asked for links to. You did the ID Quality Experiment in 2017, which kueda reported and interpreted on the results of, and included additional calculations. In which, you asked taxonomic experts to apply, so all the identifiers were considered experts. Some of keuda's main conclusions were: Accuracy is complicated and difficult to measure. What little we know suggests iNat RG observations are correctly identified at least 85% of the time. Vision suggestions are 60-80% accurate, depending on how you define “accurate,” but more like 95% if you only accept the “we’re pretty sure” suggestions.
Yet, these experiment specifically only sought taxonomic experts who had to apply and get accepted, with a total of 30 experts (a very small sample size). There were 4,336 IDs and 3,031 IDs (seemingly small sample sizes, if the experiment were to be applied to all wildlife) where the community taxon was at species level ("i.e., probably Research Grade"). As expected, the % accuracy of taxonomic experts was relatively high, but also in many ways more of a B grade (85%) than an A grade, so not that good. And the majority of those obs. either were or were inferred to be RG.
One note here is that as these past years’ posts show, the staff made more reference to distinguishing taxonomic experts from other users in the past (although, while at the same time saying IDs shouldn't be made from arguments from authority/credentials). Versus, today, the staff seldom use the term expert at all, and the eligibility bar loarie set for the validators (who are analogous to the “experts” in the past) in the most recent experiments was lowered significantly, so low that someone merely guessing a small number of IDs could quickly achieve it.
In all of the past and current experiments, the obs. and/or ID sample size, and expert/validator "sample size" (at least in the first recent experiment) have been far too low to be able to draw confident estimates from the results. Like I and other users mentioned, there are also numerous, including inherent, limitations to the design of the past and current experiments, some of which are impossible to overcome, which require caveating. kueda also noted additional such limitations or caveats to those recently made by commenters and loarie.
Overall, the large number of limitations, small obs. sample size, small validator/expert sample size (at least in some experiments), and the low accuracy findings for the CV/AI suggest that it remains very difficult to precisely estimate accuracy in general and that the validator/expert accuracy are overestimated in both past and current experiments. It would be relevant to do further research on current CV/AI accuracy, since contrary to what a commenter recently claimed there must be some correlation between CV/AI and RG accuracy. Even if we were to believe the reported 85% and 95% accuracy estimations, it can be noted that those results are very different from each other, and are mostly related to expert/validator IDs only.
2 of 2
“Anecdotally” (from experience), extensive identifiers, even ones who work in groups that may actually have 95%+ accuracy, can also intuit/infer/notice that 95% accuracy would be an overestimate as an average across all taxonomic groups. Because, as kueda said, accuracy and expertise vary significantly between groups, and we know of groups with much less review and fewer “expert” vetting. kueda and others also noted the inherent problem of groupthink, where all the “experts” think an ID is accurate but which they later learn isn’t, which would also contribute to overestimating % RG accuracy.
In conclusion, the above seems to suggest the following: the titles or descriptions of the recent and ongoing experiment results should be more significantly caveated and noted to likely be overestimates (or if not believed to be, provide compelling reason why not, and ask commenters what they think). Already, inat being estimated as 95% accurate is one of the top Google Chrome results to the keyword terms, without clarifying, so changing this post’s title would also be ideal. Also, it would be ideal to post each new experiment in three posts: the first asking for feedback on the design, the second stating how the experiment will work, and the third reporting on highly caveated results and asking the community if they interpret them to be accurate. Multiple similar iterations of the same experiments should also ideally be done: one using the current validator standard, one including all identifiers, one including the older much different “taxonomist expert” standard, etc. The sample size of identifiers, obs., and IDs should also be massively increased, and calculations on current CV/AI accuracy should be done, and possibly incorporated into the experiments, their results, and the interpretation of the accuracy of th results.
Versus, (the title itself of) “We estimate RG accuracy is 95%!,” make it sound like the estimation is highly accurate and not an overestimate. Presenting the results in a positive manner (e.g., the explanation mark) is also misleading, since 95% means 5% of all RG records sent to GBIF are wrong, and can make commenters feel like, or be perceived by others as being, “negative” to suggest reasons why the results would seem to be overestimates, incomplete, or otherwise misleading. By contrast, keuda’s earlier title and writing on loarie’s 2017 experiment was far more neutral and provided more caveats about the limitations, asked skeptical questions, etc. Overall, part of my main conclusion is that no methods yet used would even allow us to arrive at a good estimate of %RG accuracy. For this reason and the fact that many or most consider the stats overestimates, it wouldn’t be ideal to report these findings in other sources. Lastly, it also seems evident that since the staff are clearly trying to achieve and demonstrate a high % RG accuracy, that the website design should be tailored more toward ways that increase photo quality and RG accuracy.
I agree wholeheartedly.
Here's an example of how RG accuracy affects CV suggestions:
When analyzing Malva misidentifications, I found the CV suggestions to be incorrect most of the time. After correcting thousands of misidentifications, the CV suggestions are now more accurate:
https://www.inaturalist.org/projects/malva-genus-mallows-california/journal/
Using the CV-suggested species can actually propagate the problem if the data upon which the model is trained are incorrect.
So I did the Eristalis experiment again... 125 observations this time. I made it harder for the CV than I had done before by setting the taxon fliter to 'life' rather than 'by taxon', to make it more likely to get the genus wrong. (On the few occasions it tried to identify the plant or another thing in the picture, I just ignored those and picked the highest ID that seemed to have the correct target). Of the observations I could put a species on it got 86% correct this time! 16 different species on 3 continents. Those regular new releases must be making a difference. Of the 17 it got wrong, one was a species not known to the CV, 5 I noted had pictures in which the specimen was very small, some of the others were overexposed or otherwise difficult. Only 7 times I thought the CV had no excuse - including a very clear E arbustorum it bizarrely thought was Eristalinus taeniops, a decent, if slightly small picture of the densely hairy E flavipes that it thought was the very unhairy wasp Scolia dubia, and a fairly poor but still clear E tenax on a white wall that it interpreted as a cat flea! Overall, pretty brilliant.
1/4 Thanks @truthseqr. I have a few remaining thoughts following up on/reinforcing my earlier ones, that mentioned the earlier 2017 staff experiment that estimated (with many caveats and limitations) 85% accuracy. Also, the word second in the title of the current post would make readers unaware or forget that there ever was at least 3 experiments total (despite having some differences in their methods). In the 2017 experiment and current ones, a relatively small # of experienced identifiers/experts have the largest effect on the entire RG dataset accuracy (at least for groups that have experienced identifiers).
(Note: I suggest mostly now using the term “experienced identifier” instead of “expert” throughout inat, to indicate a user’s gained photographic record ID accuracy on/via inat experience, regardless of credentials or external traditional training. Real-life experts will tend to be or become experienced identifiers, anyway. Yet, I also commonly see top taxonomic experts who don’t ID on inat make misidentifications when users email them photos to ID, demonstrating that in-person specimen ID expertise and photographic record ID skill only partly overlap, and that any (incl. taxonomist) photo record identifier typically requires at least 1-2 years of regular experience to become equally as skilled for photo ID. But to clarify, I recommend all users seeking to increase ID skill also pursue and learn from traditional taxonomic training and experts, e.g., at minimum to learn to read academic ID publications for difficult taxa IDs. Anyway, to save space, I use “expert” herein to typically mean experienced photographic record identifier, and “AI” to mean CV. I still refer to validators as validators, because I don’t consider the avg. one a highly experienced identifiers given the current low validator eligibility. Lastly, I use other abbreviations and shortened sentence structures I typically don’t to shorten my comments. Finally, note that often preferred “short” comments can be misunderstood or vague given the complexity of the topic and overly-lengthy inat terminology, so it’s hard to write in fewer words.)
The small # of experienced identifiers included in the 2017 experiment is partly due to only a small # and % of taxonomic experts joining inat (when "expert" is defined the 2017 experiment way). Also, to present, these “experts" (now referring to the slightly larger but still somewhat small # of experienced inat photo identifiers) have great influential power over other identifiers when they make IDs on the same obs.; the latter typically follow expert ID advice. The fact that different year experiments mostly focus on obs. that are, were presumed to be, or are expected to become RG also makes it likely that expert users IDed a majority of the obs. Including un-doing the RG status of obs. where 2 users guessed a wrong ID in succession via using disagreeing IDs.
2/4 So, as some commenters implied, these experiments are mostly actually estimating expert ID accuracy (including the accuracy of the other users who typically "follow" the experts), and mostly for RG or to-become-RG obs. Vs., the original description of the 2024 experiments made it seem like it was testing the avg. accuracy of all obs. and/or all identifiers. Part of the expert user effect is impossible to overcome, because the community does already tend to "follow" experts, unless an experiment was to be done where each participant can't see any IDs made by other users. Regardless, one takeaway is that even if a relatively high accuracy were to be estimated, it should be clarified that that's most likely at least somewhat overestimated and is a far higher than what the avg. ID accuracy of all users would be (in all circumstances, incl. where only they made an ID), and is much higher than avg. initial AI accuracy (which was only near 50% in 2017, excepting the subset of "we're pretty sure it's this taxon" suggestions that had less-low accuracy).
As I suggested, it would seem ideal not only to a conduct number of versions of these experiments, but to intentionally consider "the" experiment and "the" eventual results of it as including or considering a complex of different iterations of the experiments in presenting and interpreting them and asking if users/readers agree. For e.g., the following experiments would be useful to conduct (possibly multiple times), and it's also necessary to massively increase the obs. sample size for each (and we already have data from small obs. sample size experiments, which we can keep in consideration when later interpreting all results:
The current experiment version, the current experiment where all users (who participate) are incl., the current experiment with a more difficult validator eligibility, the current experiment with a much higher validator eligibility (and/or repeat the similar 2017 experiment), an experiment where validators can't see what IDs other users made or at least can't see who the users are, a current assessment of initial AI accuracy (when an observer uses AI for the initial obs. ID), an assessment of AI accuracy when an AI ID by a user after a preceding user’s AI-ID was incorrect, an assessment of AI accuracy when an AI ID by a user after the preceding user’s AI-ID was correct but was only at a broad taxon rank, and iterations of all of these experiments. Add’l variations or iterations should also try designs where most (or varying percents) of incl. obs. are expected to become RG or casual, or (in separate versions) Needs ID or casual by the experiment’s conclusion.
3/4 Another thing to assess is how many misIDs are made on obs. that later turn out to get a corrected. Naturally, all this would require many experiments, but the matter is actually so inherently complex that to draw meaningful, accurate conclusions from them might require that, possibly even conducting experiments over years, and then making more accurate interpretations of findings by considering all the results from all the experiments/studies.
And by remembering and incorporating into the results interpretation that many design limitations and caveats (some unavoidable) would remain. And, given the complexity of what these experiments try to assess, it also seems likely that any result (even if conducting and repeating the many experiment types) would be at least somewhat imprecise and require caveating and interpretative commentary, and most likely be at least slight overestimates (for reasons given). Any AI accuracy assessment experiments would also be limited since the Seek mobile app doesn’t take into account Nearby, and that the desktop inat version of the AI and Compare/Suggestions differ (suggest different IDs).
Vs., merely reporting experiment results like the recent one w/o caveats can be very misleading. Relatedly, the title of the recent experiment result post may misleadingly sound like it included all or the avg. of all identifiers IDs and/or incl. all inat obs. The title reference only to RG could also make it seem like it assessed the accuracy of inat obs. of every status. I predict that if the findings from the recently reported 2024 experiment were submitted to an academic journal, that at least if reviewers/editor were aware of how inat works, they’d note many of these and add’l limitations, caveats, uncertainty in results, and need for add’l experiments, greater obs. sample sizes, etc. In the event those (or future) experiment findings were submitted to a journal or shared in a magazine, presentation, podcast, etc. (which typically have less methods/results vetting than journals), the reviewers/editors should be made aware of the 2017 experiment and kueda’s comments on it, the full list of current limitations and caveats, etc., incl. determining whether the small obs. sample size would make any even relatively precise accuracy estimation impossible.
Otherwise, the results could seem to imply that (all-taxa) inat RG accuracy is high and higher than for sites like Bug Guide, which seems unlikely, since most BG IDs are made by users BG designates as “experts” and are restricted to a smaller geographic and taxonomic scope). Also, the inat AI (especially initial AI IDs) accuracy remaining so low, at least in some somewhat-to-very cryptic groups, is an indication that avg. user ID accuracy (in circumstances when they make the first ID) is far lower than the recent accuracy estimations that actually mostly directly or indirectly pertain to inat expert IDs being made on the same obs. And/or (depending on how the AI currently works), it could indicate that all avg.-user regular IDs or their non-initial-AI IDs have low accuracy.
4/4 The following e.g.’s can be considered further/used. A small # of experts recently completed IDing all 6,000+ global Delta obs. (somewhat cryptic). But, the avg. identifier Delta IDs and initial-AI-IDs of Needs ID Delta obs. remain similarly inaccurate (no noticeable difference).
I and add’l experts also IDed approx. all European Eumeninae (very cryptic overall) Needs ID obs. Mostly reviewing Needs ID obs., I found/corrected 30 to 50% obs. that had been misidentified to genus by users or the AI. Yet, the current AI accuracy there still remains similarly low, even for genus suggestions. This also means that if the experts (and to clarify, anyone can in principle become experienced, it isn’t a label necessarily excluding anyone) were to suddenly stop IDing, the low AI accuracy and avg. user ID obs. ID accuracy would further lower overall inat % accuracy. This means the AI at least at least currently is at least partly counteracting wrt Needs ID and RG inat obs. accuracy.
Add’l AI problems contribute, e.g., that the Compare/Suggestions Visually Similar filter seems not to incorporate the Location filter. Furthermore, the fact that even locations with faunas currently completely accurately IDed by experts would regress to lower user and AI accuracy if the experts left, indicates that multiple add’l aspects of inat are currently at least partly counteractive wrt accuracy. It’s unfortunate that the hard-won complete ID of regional faunas could be entirely lost in such a scenario, esp. because that would mean the avg. remaining regular identifier would also be ignoring the available past obs. and comment history explaining how to ID the complete fauna experts “left behind for them.” And, no user including experts can ID forever, and even active high-volume experts eventually can’t sustain their volume (incl. me, in the future), so the scenario is possible.
I’m unable to put as much time writing/thinking about this in this post or future ones, so ask people to mostly refer back to my comments here and elsewhere, or they can ask me in the posts specific questions/comments. In future inat accuracy posts, I’ll also make briefer comments if any, but may still actually disagree with some design methods, results, and results interpretations/portrayals even if I don’t comment, if they contradict or overlook points I and others made. Last thoughts. Re: muir suggesting this post be put on the forum for moderation tools etc., I instead agree with the curator who noted why it should be here so everyone can comment (many experts don’t even use the forum). Ironically, a forum mod did flag one of my above comments, claiming I insulted Scott, but Scott resolved that flag as that I didn’t, so imo and demonstrably ime forum (and sometimes inat) moderation is sometimes unfair or inaccurate. Finally again, I consider the current Google search term top result reference to inat estimated to be 95% accurate too misleading, since many Google users won’t read the details past the title, so suggest editing the post title.
Thanks everyone for another very successful experiment. We're learning a ton. We added a Results section to this post.
These estimates of avg. accuracy of the entire RG obs. dataset are in line with our expectations, largely because the iNat. dataset is skewed towards a relatively small number of common, easy to ID species (e.g. mallards, monarchs, etc.) that have an outsized impact on the average.
A. mellifera would be another easy species e.g., with verifiable RG obs. accounting for 26% of all verifiable Bees obs. This in combination w/ the fact that community and individual IDs are mostly by or influenced by experienced identifiers (“experts”), who can ID nearly 100% of the easy species in their groups, means that the results are mostly only about the accuracy of experts and those following them IDing the most common species that (increasingly over time as obs. are added) flood out all other species. Attesting to how "easy" [verifiable obs. only] such species are for experts to ID is overall, I found in Jan. that of the total of all verifiable obs. all global genus Apis species, 99.22% were RG and presumed correctly IDed, vs. only 58% of the total global verifiable Bees (all species) obs. that have been IDed so far were RG (species= 3,511).
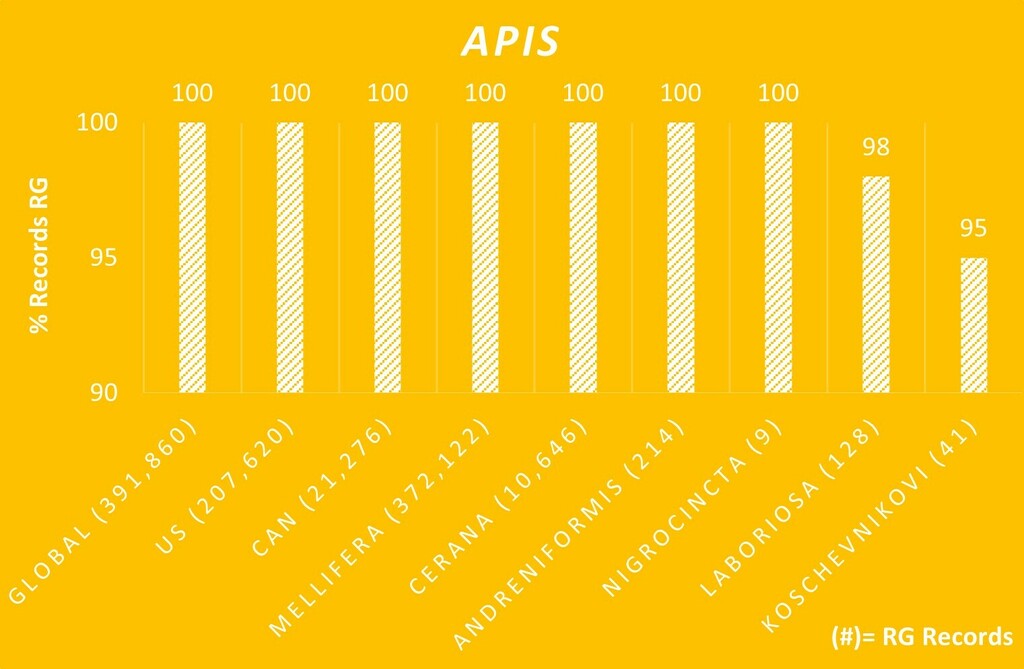
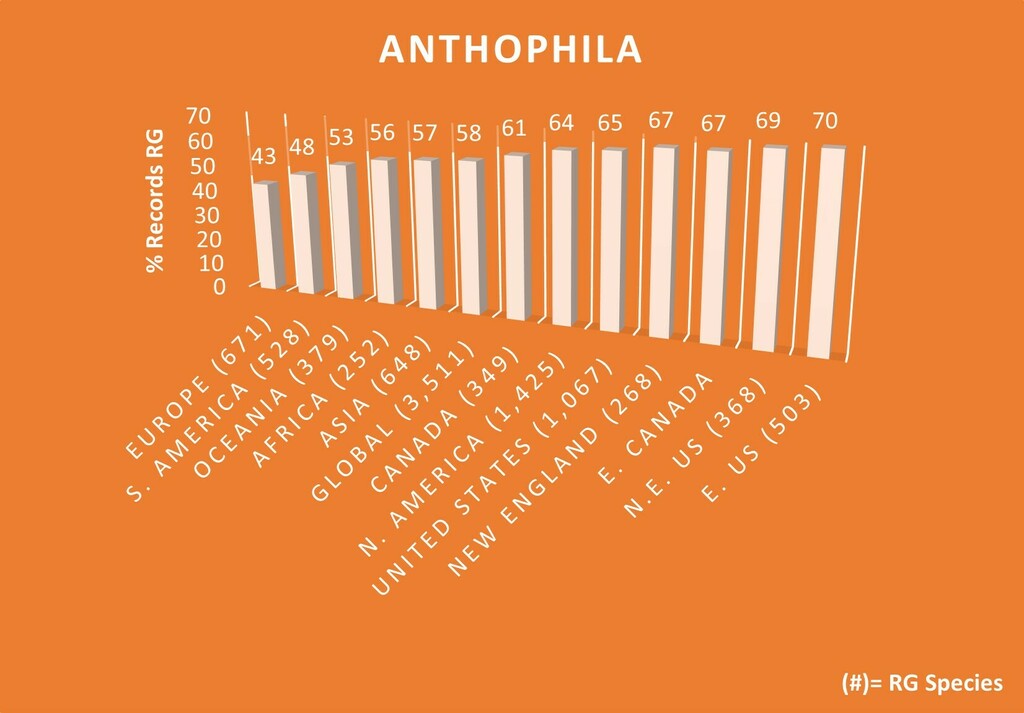
Furthermore, global bee species overall range from easy, slightly, highly, to impossibly difficult to ID to species (1,444 verifiable RG global bee spp. have been IDed to date). I'm also unsure if broader-than-species taxa that nonetheless also flood out most other bee species that are easy to make RG via DQA (and likewise for other taxa groups) (e.g. Zadontomerus and Dialictus) factored into this experiment. If the caveats and explanations were made clearer in the result designs, results, and the results titles/reporting, I may agree that these specific experiments are somewhat accurate but possibly still insufficiently imprecise due to obs. and/or identifier sample sizes (which the designer and many others stated might be a limitation). So, these results can still be potentially misleading or mostly involving "atypical" circumstances that aren't made abundantly clear upfront to readers (incl. e.g. in top Google search results). The fact that the RG accuracy results are nearly 100% reinforces this, which is also nearly the same percent of the verifiable A. mellifera obs. that are RG at any given time in my calculations.
While experiments like this can be useful for showing what they can based on their specific design, to truly report on what many would consider a more accurate, common sense, or complete accounting of inat accuracy would require doing more experiments, including ones that exclude all the numerous easy species (note: this is actually done in some publications, although methods vary) or in some way to include equal numbers of obs. per species, and to use larger sample sizes (etc., see previous comments). Because, for e.g., we know most observers observe the most abundant and easy to ID species in urban areas (a bias that was also scarcely if at all mentioned), which flood out most of the other site-wide species data. It is good that many users participated, though, and I'm interested to see what additional experiments or ones that take participant feedback upfront and post-experiment interpretations into account may find. I still predict BG has hgiher accuray though, for e.g.
Thanks for the update! If I may make a small suggestion to the methods, I would suggest running the experiment such that it includes at least one weekend day. I assume all the validators are just volunteers and depending on how busy things get at work more may respond if the validation time included some non-work days. I saw the response rate this time was 64% vs. 72% for the first experiment.
Great that these experiments are continuing!
I just want to echo a couple of people above in saying that
it would be great to have this conversation on the forum instead where we can respond more easily to other people's comments, reference in other forum threads, etc.
Also...to echo someone else's comment that I think it can be a bit misleading to use blanket statements such as the "research grade accuracy is 97%" subtitled on the main page. This is a complex topic and as touched on above, the exact meaning of accuracy in this context is slightly ambiguous... varies massively by taxon, etc. Clipping the number into a headline like that undermines things a little in my opinion, however interesting the data and experiment might be. Soundbite numbers tend to be endlessly repeated on the forum as well as off site as hard data, usually lacking context, even though at least with the last study which I looked into in more depth, it seemed the 95% number which was being touted lacked real substance. To me the numbers here seem again problematic to float around without context, however much insight they might provide to everyone. Not that it's not good to report the findings, but yes... good to not fall into the trappings of a sort of tabloid headline science approach.
Those thoughts aside, Great that there is a responsiveness from the staff to community feedback and that they are being developed further. I hope they continue to be a regular feature and I really hope that alternative methodologies are also explored.
It would be great to hear discussion about the approach more generally on the forum.
Thank you very much for the effort to optimize methods to improve the identification of the species registered in iNat, also in a community way, which is a great challenge. This results in better quality of information available to everyone.
Greetings!
I agree with most of @sbushes comment, except that this discussion should be had instead on the forum. One additional reason the discussion should be had here on inaturalist is that the experiment is attempting to recruit users to participate, by using direct private messages. (Side note: private messages between users aren't actually currently private to the staff, only their private messages to each other are, which is wrong, since the staff should only need to see private user chats that get flagged.) So, that would be like if in a next experiment, inat sends private messages to inat users, but then tells them that the discussion will be on the forum, but only a portion of the users even have a forum account, and many experienced identifiers in particular don't use the forum.
It would also be confusing if the expertiments were discussed both on inat and on the inat forum, although I suppose non-staff forum users could choose to start additional forum discussions on these, but which would exclude some of the participants. As for following the various "threads" on this post, it actually isn't that confusing or hard to follow. This is exactly as the Bug Guide forums work and they have no moderation at all. As for me, I've given long detailed thoughts on the 2017 experiment most users seemed unaware of and on these two recent 2024 ones, and feedback for what I think would improve them, and things to avoid. So, I won't be repeating anymore long messages like that for subsequent experiments; if anything I'd only write briefer comments or otherwise recommend users refer back to my earlier comments.
One last slightly problematic issue comes to mind: A. mellifera (western honeybee) is one of the most observed and easiest to ID species, but in many locations are from managed hives/apiaries but not annotated as captive. There's a general lack of understanding among many users that if the bees are literally seen coming from a managed bee container, they should be categorized as captive. It's even come up in the past that evidently some staff think it's okay to keep honeybees known to be from managed colonies as wild, which disagrees with the DQA and guidelines.
The slight problems I'm pointing out here is that a large portion of the species included aren't actually wild, and therefore should've been excluded (which affects sample size, etc.). I suggest that the guidelines or other guidance add clearer guidance on Apis mellifera: if they're known to be managed they should be categorized as captive (making them casual grade) but if one is seen in the wild where both wild and managed honeybees can occur they can be left categorized as wild.
My understanding is that this is just a measure of identification accuracy, and that captive/cultivated observations were included. While it would be good to improve the accuracy of captive/cultivated labeling, it doesn't directly intersect with the goals of this experiment. As long as the ID is correct, it doesn't affect the results in any way I can think of if it's erroneously labeled wild.
Was disappointed to not be invited to this round but hope to be in a future one!
I'm not sure the numbers are meaningful without more context. An accuracy value for iNaturalist as a whole does not tell you what you want to know unless the sampling biases in your data usage match the sampling biases in the data used to create the accuracy value.
I think it's pretty obvious that there are subsets of the iNaturalist data where ID accuracy is, for all practical purposes, 100%. And subsets where ID accuracy is more like 25-30%. As a data user, I want to know where I am relative to that variation.
I strongly agree with @matthias22 that you should only ask people to identify species on a much smaller scale than that of a continent. Many of us botanists are experts on a certain place, and know what all the taxa are in a well-defined area, such as Anza Borrego Desert State Park, San Jacinto Mountain, San Gabriel Mountains, etc. When asked to identify something in Texas or Mexico, even though that is the same continent, we are handicapped since we don't know what additional look-alike species may be present there. You really should only ask people to identify observations "as an expert" in areas where they actually are experts.
Thank you for your invitation. Pity, your time window was much too small to take part in this interesting event.
Just to mention: It was a great fun and honour for me to be part of the experiment!
I second @matthias22 about narrowing down to state or other localized geographic region. As somebody who identifies things almost exclusively in one or two states (or even counties), I did my best on these experiments, but felt under-qualified due to the fact that many observations I was being asked to ID were hundreds or thousands of miles away from my knowledge-base and resources. Thanks for the opportunities and continuing to refine the platform and its use and accuracy.
I would like to be offered the 'thousand obs' in a project - then we could ID by preferred taxon and location.
Would be interesting to see the result from doing it that way around.
From this batch there were a few invasive / garden plants I recognise - for example.
Intriguing idea, @dianastuder! One could think of an experiment where only the observations are chosen but not the validators. The test observations would be made public and the whole community invited to weigh in according to each one's expertise. Each identifier could pick the taxa and regions they are familiar with. This might be a good system for validating rare species with relatively few observations that require fairly specialized expertise. Definitely not a good method for easily recognizable taxa with tens of thousands of observations such as mallards and honey bees. I don't know whether this is feasible. Perhaps it's just a crazy idea but I wanted to put it out there. It has a crowd aspect to it where the community could work together in order get an observation to the finest taxonomic level possible and to the greatest ID accuracy possible.
I would definitely like to see future experiments that focus on more circumscribed taxa or regions. Doing these experiments for every kingdom worldwide is very generalized. It would be helpful to our various communities such as botanists, entomologists. herpetologists, etc. to have more specific experiments that provide more applicable results to their fields of study.
Definitely love all this, and I am very motivated to contribute to the best of my abilities to future experiments!
Agreeing with additional comments, as I also suggested it would be useful to conduct multiple different kinds of experiments, and repeat them. Both to get a better understanding of "overall accuracy" (but I think there should be multiple versions of that too, e.g., one that excludes the easy abundant species), and also to gain findings about particular specialized experiments in their own right. In other words, that there are multiple meaningful measures of accuracy, both when considered together and in their own right, and that by conducting them the results will reciprocally increasingly inform how to interpret the accuracy and results of each of the accuracy experiments and of all of the experiments considered as one experiment ("complex of experiments").
As this comment implies, it is somewhat in keeping with the goals of the original design of the 2017 and two 2024 experiments, there also remains a need to attempt to understand an average, overall, common sense measure of "overall accuracy," but how we define that matters (e.g. only using experiment designs that are flooded with mallards and honeybees, vs. conducting those plus additional experiments excluding the abundant species, plus drawing inferences from specialized studies to help interpret the "average accuracy"). Becuase as time goes on, there will only be an increasing number and proportion of abundant urban species like mallards and honeybees, making some of the experiments as designed so far remain near 100%, which is an indication that something more needs to be modified in the design(s).
Although, the "100% accuracy-results" experiments can also be included as one among multiple sources of information in understanding accuracy, so weren't entirely useless, but just don't teach us very much yet on their own and it seems inaccurate to report their results in public venues, unless significantly caveating that the limitations may mean the estimation is both unknown, an overestimate, and possibly very far off. Observation (and identifier) sample size (and varying them, even among iterations of each type of experiment) also matters. Finally, as mentioned, studying the average accuracy of non-RG obs. (Needs ID), initial-AI accuracy, AI-accuracy after someone's made a human or AI ID, etc., will also be useful and necessary, for those truly seeking to understand accuracy in a more accurate and meaningful way. Which may take a long time, but would be worthwhile.
@tonyrebelo and @cthawley concerns if seeing existing IDs is not a blind assessment are true, but that is the "living" situation in IDing observations as well as in both experiments at present. But this is a chance to prove your own knowledge whether you can "agree" to already present IDs or if you have to look up other sources before giving an ID. In case of your own uncertainty or doubts, not make any ID, leave the present page and move on to the next observation.
However it would be a nice and important experiment to make additionally a blind one, once. iNat would be able to measure validator skills and you as an IDer could learn as well a lot.
Obviously, the fact that ID are not "blind" is less than optimum. However, I would think it's less of a problem with this experiment than in most of iNaturalist. We want to be careful and get it right because we know our ID will be reviewed by others.
Good discussion. In a conventional research setting (e.g., taxonomists working at a natural history museum or a university), identifications are not blind in many cases, either, unless one deals with newly collected specimens. The determination history of specimens in natural history collections is carefully recorded and visible to any researcher who works on those specimens. In this regard, iNaturalist as a citizen platform is no different. Observations first come in as unidentified and then the determination history is carefully documented and visible to all the citizen scientists who look at them later. Therefore, I would not overemphasize this issue. Citizen science and institutional science work under very similar conditions in this regard. That does not mean, of course, that it wouldn't be interesting to find out how much ID choices are influenced by previous IDs. In fact, that's a fascinating question which I personally find intriguing. Based on my own observations here on iNaturalist, previous IDs can have a great influence on subsequent IDs, including when they are wrong. Initial incorrect IDs sometimes even influence experienced identifiers who know better to agree with them. The "Agree" button is deceptively easy to use :) Research on this is definitely to be encouraged. I would love to see a study where the same observation is presented to two groups of validators, one with a correct ID and one with an incorrect initial ID. And then quantify the different outcomes. That would obviously not be possible with existing sets of observations because one would have to create duplicates.
One difference between iNat and conventional research is the workload, A Protea taxonomist in the Cape would expect no more than 20 specimens per week, whereas the daily amount on iNaturalist (798 the previous week for s Africa) is well above that. In addition, on the CNC and GSB thousands of identifications (e.g 3,118 Proteaceae for GSB2023 in s Africa) may be expected over a week: it is literally glance and ID - where an initial wrong ID can totally blind one. I have several times realized the error 3-4 identifications on, and had to backtrack to correct the ID. And also been totally puzzled a few weeks later by an ID made during these events (e.g. could the ID have been intended for a previous observation?, or a misclick on the options provided?).
The situation is totally different in these accuracy experiments, where the care and time more typical of a herbarium/museum specimen will be spent making an ID.
To my mind the big difference is the degree of checking. Hardly anyone will check my Proteaceae herbarium IDs for decades, but the iNat ones will be scrutinized by a dozen interested people within the week (versus a day for any bird IDs), most of whom dont comment or add an ID, unless something is blatantly wrong (or an exciting observation). So one has to be extra careful with specimens - for observations on iNataralist being wrong is more about the embarrassment than the legacy.
Well put, @tonyrebelo !
Suggestion for selecting experts: each species has a list of top identifiers. Why not choose your experts from that list?
@dianastuder, I tried your method of marking and retrieving duplicates. I find it time-consuming & not very effective. I've resorted to keeping track of duplicates in an Excel spreadsheet.
time-consuming & not very effective
Both true. But mine is visible to future identifiers. Your spreadsheet is only for your benefit.
I hope for more new DQA.
Duplicates (one, or more, to be deleted). Meanwhile Casual to take them away from identifier's backlog.
One subject scattered across many obs (to be combined). Ditto re Casual.
If the issue would only affect me - I would Mark as Reviewed - and keep moving along.
@xris has created a field: https://www.inaturalist.org/observation_fields/15623
Observation field:
Duplicates ObservationFull URL of the initial observation, which this observation duplicates, of the same instance of an organism, e.g. when there are multiple observers during a bioblitz.
In the ID curation tool, just click the Annotations tab, add the field "Duplicates Observation" and enter the url of the original (the first observation in the set by date:time added).
Or, on the observation, in the Observation Fields panel - do the same.
You can easily access the duplicates here:
https://www.inaturalist.org/observation_fields/15623
or here:
https://www.inaturalist.org/observations?verifiable=any&place_id=any&field:Duplicates%20Observation
(and you can apply further filters as required).
Personally, this is far quicker than using Excel, and is available to anyone interested.
Thanks, @tonyrebelo. I'm aware of that field and I was using it for awhile. The part I was missing was how to find the duplicate observations after using that field. Thanks for the URLs. That helps a lot. I'll update the duplicates I've found.
@tonyrebelo I've been using that field, and I totally forgot I had created it! It came from my work during City Nature Challenges, for which I've been a local "captain" for several years.
Thanks @xris - that'a a great solution to the duplicates problem.
Is it possible change the iNat blog display so we can search for our own comments?
If I search my name I can find the @mentions - then have to scroll up to hunt down what people are responding to.
Posted by truthseqr - IS searchable for me.
But my own comments say - Posted by you - and searching for you ...
@truthseqr Your "Suggestion for selecting experts: each species has a list of top identifiers..."
True, but if iNat would only select the first ones, you may end up with a surprise and a very skewed result. I will now only reflect of top-identifiers of lepidoptera species, a field I am familiar with and having contributed a small share of IDs as a lay-identifier. If this list would reflect only those identifiers who contributed mostly "improving" IDs, or the first "supporting" or "leading" flagged ones (the latter in case of correcting previous wrong IDs), then this assumption would be very good.
Matter of fact that is a minority. While otherwise very knowledgeable experts often are found in the back ranks, pushed back by multi-clickers with "supporting" percentages above 85%. OK each of us can decide whom to contact to be helped with difficult IDs. An automated system that does only select the top-notch identifiers will not reflect the real expertise in iNat, hence a random selection is still the best.
Add a Comment